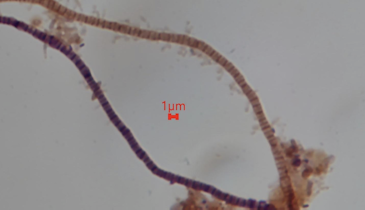
Wastewater treatment plant operators have relied on a variety of monitoring tools — DO, pH, SV30/SVI, sludge depth — to keep facilities running efficiently and within permit requirements. While microscopic exams and ATP measurements provide insight into the health of the biological...
Using Molecular Diagnostics for Wastewater Monitoring
Popular Stories
Discussion
Comments on this site are submitted by users and are not endorsed by nor do they reflect the views or opinions of COLE Publishing, Inc. Comments are moderated before being posted.